Study is first to show role of genomic changes in specific braincells in Alzheimer'sdisease uniofexeter NatureComms
). To identify DNA methylation sites associated with AD neuropathology we conducted an
in which DNA methylation at each probe was regressed against the three measures of tau and amyloid pathology using mixed effect regression models where age, sex, experimental batch, PC1 and derived brain cell proportions were included as fixed effects and individual was included as a random effect.
for each of the five neuropathology measures separately using the same set of covariates. In addition, we ran analyses where cell proportions were regressed against neuropathology in each brain region using linear regression models, controlling for age and sex. To identify tissue-specific effects, linear regression models were run in each brain region for the three main AD neuropathology measures controlling for age, sex, experimental batch, PC1, and derived cell proportions.
results were subsequently processed using the
Danmark Seneste Nyt, Danmark Overskrifter
Similar News:Du kan også læse nyheder, der ligner denne, som vi har indsamlet fra andre nyhedskilder.
 Single-cell multi-omics profiling links dynamic DNA methylation to cell fate decisions during mouse early organogenesis - Genome BiologyBackground Perturbation of DNA methyltransferases (DNMTs) and of the active DNA demethylation pathway via ten-eleven translocation (TET) methylcytosine dioxygenases results in severe developmental defects and embryonic lethality. Dynamic control of DNA methylation is therefore vital for embryogenesis, yet the underlying mechanisms remain poorly understood. Results Here we report a single-cell transcriptomic atlas from Dnmt and Tet mutant mouse embryos during early organogenesis. We show that both the maintenance and de novo methyltransferase enzymes are dispensable for the formation of all major cell types at E8.5. However, DNA methyltransferases are required for silencing of prior or alternative cell fates such as pluripotency and extraembryonic programmes. Deletion of all three TET enzymes produces substantial lineage biases, in particular, a failure to generate primitive erythrocytes. Single-cell multi-omics profiling moreover reveals that this is linked to a failure to demethylate distal regulatory elements in Tet triple-knockout embryos. Conclusions This study provides a detailed analysis of the effects of perturbing DNA methylation on mouse organogenesis at a whole organism scale and affords new insights into the regulatory mechanisms of cell fate decisions.
Single-cell multi-omics profiling links dynamic DNA methylation to cell fate decisions during mouse early organogenesis - Genome BiologyBackground Perturbation of DNA methyltransferases (DNMTs) and of the active DNA demethylation pathway via ten-eleven translocation (TET) methylcytosine dioxygenases results in severe developmental defects and embryonic lethality. Dynamic control of DNA methylation is therefore vital for embryogenesis, yet the underlying mechanisms remain poorly understood. Results Here we report a single-cell transcriptomic atlas from Dnmt and Tet mutant mouse embryos during early organogenesis. We show that both the maintenance and de novo methyltransferase enzymes are dispensable for the formation of all major cell types at E8.5. However, DNA methyltransferases are required for silencing of prior or alternative cell fates such as pluripotency and extraembryonic programmes. Deletion of all three TET enzymes produces substantial lineage biases, in particular, a failure to generate primitive erythrocytes. Single-cell multi-omics profiling moreover reveals that this is linked to a failure to demethylate distal regulatory elements in Tet triple-knockout embryos. Conclusions This study provides a detailed analysis of the effects of perturbing DNA methylation on mouse organogenesis at a whole organism scale and affords new insights into the regulatory mechanisms of cell fate decisions.
Læs mere »
 Palate-Cleansing Fashion Is On The Menu Next SeasonOn the spring/summer 2023 ready-to-wear catwalks at New York, London and Milan Fashion Weeks, the fashion was driven by design DNA. Essence-driven design that acts a palate-cleanser to the overwrought and the extraneously embellished.
Palate-Cleansing Fashion Is On The Menu Next SeasonOn the spring/summer 2023 ready-to-wear catwalks at New York, London and Milan Fashion Weeks, the fashion was driven by design DNA. Essence-driven design that acts a palate-cleanser to the overwrought and the extraneously embellished.
Læs mere »
 Single-cell multi-omics profiling links dynamic DNA methylation to cell fate decisions during mouse early organogenesis - Genome BiologyBackground Perturbation of DNA methyltransferases (DNMTs) and of the active DNA demethylation pathway via ten-eleven translocation (TET) methylcytosine dioxygenases results in severe developmental defects and embryonic lethality. Dynamic control of DNA methylation is therefore vital for embryogenesis, yet the underlying mechanisms remain poorly understood. Results Here we report a single-cell transcriptomic atlas from Dnmt and Tet mutant mouse embryos during early organogenesis. We show that both the maintenance and de novo methyltransferase enzymes are dispensable for the formation of all major cell types at E8.5. However, DNA methyltransferases are required for silencing of prior or alternative cell fates such as pluripotency and extraembryonic programmes. Deletion of all three TET enzymes produces substantial lineage biases, in particular, a failure to generate primitive erythrocytes. Single-cell multi-omics profiling moreover reveals that this is linked to a failure to demethylate distal regulatory elements in Tet triple-knockout embryos. Conclusions This study provides a detailed analysis of the effects of perturbing DNA methylation on mouse organogenesis at a whole organism scale and affords new insights into the regulatory mechanisms of cell fate decisions.
Single-cell multi-omics profiling links dynamic DNA methylation to cell fate decisions during mouse early organogenesis - Genome BiologyBackground Perturbation of DNA methyltransferases (DNMTs) and of the active DNA demethylation pathway via ten-eleven translocation (TET) methylcytosine dioxygenases results in severe developmental defects and embryonic lethality. Dynamic control of DNA methylation is therefore vital for embryogenesis, yet the underlying mechanisms remain poorly understood. Results Here we report a single-cell transcriptomic atlas from Dnmt and Tet mutant mouse embryos during early organogenesis. We show that both the maintenance and de novo methyltransferase enzymes are dispensable for the formation of all major cell types at E8.5. However, DNA methyltransferases are required for silencing of prior or alternative cell fates such as pluripotency and extraembryonic programmes. Deletion of all three TET enzymes produces substantial lineage biases, in particular, a failure to generate primitive erythrocytes. Single-cell multi-omics profiling moreover reveals that this is linked to a failure to demethylate distal regulatory elements in Tet triple-knockout embryos. Conclusions This study provides a detailed analysis of the effects of perturbing DNA methylation on mouse organogenesis at a whole organism scale and affords new insights into the regulatory mechanisms of cell fate decisions.
Læs mere »
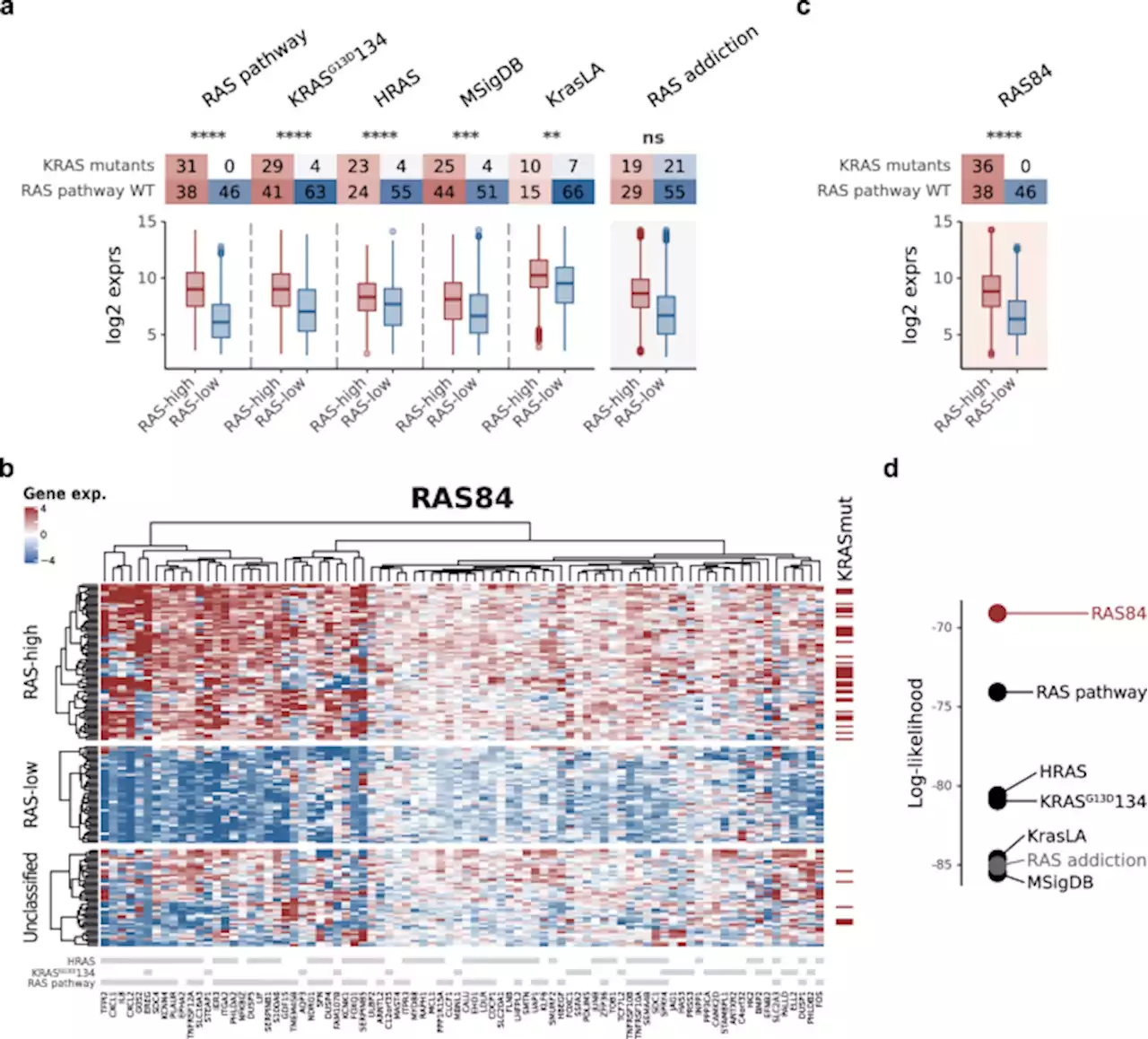 RAS oncogenic activity predicts response to chemotherapy and outcome in lung adenocarcinoma - Nature CommunicationsMutations in RAS oncogenes and related pathways are frequent in lung cancers. Here, the authors derive a RAS gene expression signature and a machine learning classifier to predict drug response and clinical outcomes in lung adenocarcinoma and other solid tumours, with improved performance over KRAS mutations alone.
RAS oncogenic activity predicts response to chemotherapy and outcome in lung adenocarcinoma - Nature CommunicationsMutations in RAS oncogenes and related pathways are frequent in lung cancers. Here, the authors derive a RAS gene expression signature and a machine learning classifier to predict drug response and clinical outcomes in lung adenocarcinoma and other solid tumours, with improved performance over KRAS mutations alone.
Læs mere »
